Desktop Survival Guide
by Graham Williams


|
DATA MINING
Desktop Survival Guide by Graham Williams |

|
|||
Random Survival Forests |
Example 1: Veteran's Administration lung cancer trial from Kalbfleisch and Prentice. Randomized trial of two treatment regimens for lung cancer. Minimal argument call. Print results, then plot error rate and importance values.
> library(randomSurvivalForest) |
randomSurvivalForest 3.6.2 Type rsf.news() to see new features, changes, and bug fixes. |
> data(veteran, package = "randomSurvivalForest") > veteran.out <- rsf(Survrsf(time, status)~., data = veteran) > print(veteran.out) |
Call:
rsf.default(formula = Survrsf(time, status) ~ ., data = veteran)
Sample size: 137
Number of deaths: 128
Number of trees: 1000
Minimum terminal node size: 3
Average no. of terminal nodes: 21.38
No. of variables tried at each split: 2
Total no. of variables: 6
Splitting rule: logrank
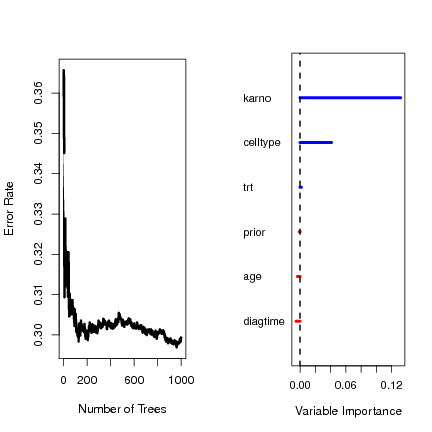
Estimate of error rate: 29.53%
|
> plot(veteran.out) |
Importance Relative Imp
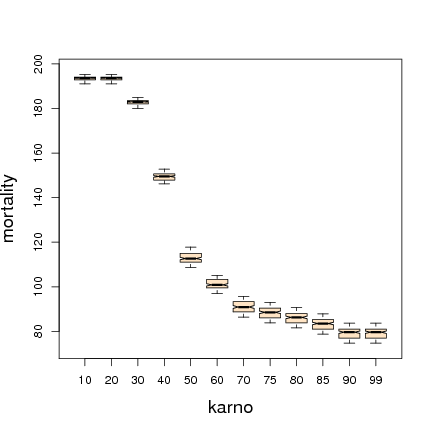
karno 0.1386 1.0000
celltype 0.0363 0.2618
diagtime 0.0045 0.0326
prior 0.0016 0.0114
trt 0.0003 0.0024
age -0.0027 -0.0196
|
Example 2: Richer argument call (veteran data). Forest is saved by setting 'forest' option to true (see 'rsf.predict' for more details about prediction). Coerce variable 'celltype' as a factor, and karnofsky score as an ordered factor to illustrate factor useage in RSF. Use random splitting with 'nsplit'. Use 'varUsed' option.
> data(veteran, package = "randomSurvivalForest")
> veteran.f <- as.formula(Survrsf(time, status)~.)
> veteran$celltype <- factor(veteran$celltype,
labels=c("squamous", "smallcell", "adeno", "large"))
> veteran$karno <- factor(veteran$karno, ordered = TRUE)
> ntree <- 200
> mtry <- 2
> nodesize <- 3
> splitrule <- "logrank"
> nsplit <- 10
> varUsed <- "by.tree"
> forest <- TRUE
> proximity <- TRUE
> do.trace <- 1
> veteran2.out <- rsf(veteran.f, veteran, ntree,
mtry, nodesize, splitrule, nsplit,
varUsed = varUsed, forest = forest,
proximity = proximity, do.trace = do.trace)
> print(veteran2.out)
|
Call:
rsf.default(formula = veteran.f, data = veteran, ntree = ntree, mtry = mtry, nodesize = nodesize, splitrule = splitrule, nsplit = nsplit, forest = forest, proximity = proximity, varUsed = varUsed, do.trace = do.trace)
Sample size: 137
Number of deaths: 128
Number of trees: 200
Minimum terminal node size: 3
Average no. of terminal nodes: 21.385
No. of variables tried at each split: 2
Total no. of variables: 6
Splitting rule: logrank *random*
Number of random split points: 10
Estimate of error rate: 29.67%
|
> plot.proximity(veteran2.out) |
Take a peek at the forest ...
> head(veteran2.out$forest$nativeArray) |
treeID nodeID parmID contPT mwcpSZ 1 1 1 3 4 0 2 1 1 2 NA 1 3 1 1 0 NA 0 4 1 3 5 54 0 5 1 3 5 42 0 6 1 3 0 NA 0 |
Average number of times a variable was split on.
> apply(veteran2.out$varUsed,2,mean) |
trt celltype karno diagtime age prior
1.775 3.240 4.810 4.295 5.200 1.065
|
Partial plot of top variable.
> plot.variable(veteran2.out, partial = TRUE, npred=1) |